The Drosophila melanogaster suppressor of sable gene, su(s), encodes a novel, 150-kDa nuclear RNA binding protein, SU(S), that negatively regulates RNA accumulation from mutant alleles of different genes which have transposon insertions within the 5′ transcribed area.
On this examine, we delineated the RNA binding area of SU(S) and evaluated its relevance to SU(S) operate in vivo.
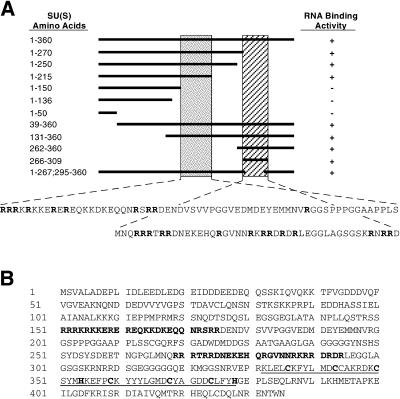
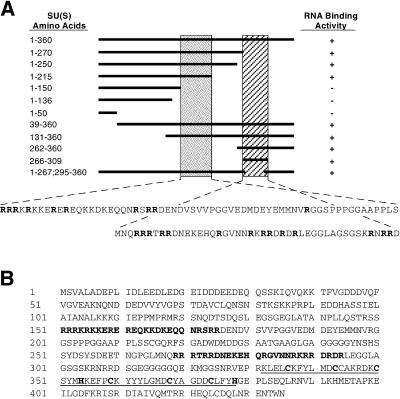
Because of this, we’ve got outlined two arginine-rich motifs (ARM1 and ARM2) that mediate the RNA binding exercise of SU(S). ARM1 is required for in vitro high-affinity binding of SU(S) to small RNAs that have been beforehand remoted by SELEX (binding website choice assay) and that include a typical consensus sequence.
ARM1 can also be required for the affiliation of SU(S) with larval polytene chromosomes in vivo. ARM2 promotes binding of SU(S) to SELEX RNAs that lack the consensus sequence and apparently is neither crucial nor adequate for the steady polytene chromosome affiliation of SU(S).
Use of the GAL4/UAS system to drive ectopic expression of su(s) cDNA transgenes revealed two beforehand unknown properties of SU(S). First, overexpression of SU(S) is deadly. Second, SU(S) negatively regulates expression of su(s) intronless cDNA transgenes, and the ARMs are required for this impact.
Contemplating these and former outcomes, we suggest that SU(S) binds to the 5′ area of nascent transcripts and inhibits RNA manufacturing in a way that may be overcome by splicing complicated meeting.
Localization of regulatory protein binding websites within the proximal area of human myometrial connexin 43 gene.
Parturition is preceded by a big enhance in hole junctions between myometrial clean muscle cells. Connexin 43 is the most important structural protein of myometrial hole junctions.
To discover transcriptional regulation of the myometrial Cx43 gene, we used DNase I footprinting, electrophoretic mobility shift and transient transfection assays to look at a 312 bp promoter area (-164 to +148) of the gene, using human myometrial cell cultures and nuclear extracts.
The DNase I research confirmed 4 areas of nucleoprotein interactions. Safety of area 1 (-80 to -31) encompassed an Activator Protein 1 (AP1) (-44 to -36) and two Specificity Protein 1 (Sp1) (-77 to -69 and -59 to -48) consensus sequences. Areas 2 to four included the transcription initiation website (-10 to +25), an Ets/NF-kB consensus sequence (+47 to +74) and a TA-rich area (+81 to +101) respectively.
Gel mobility shift and supershift assays demonstrated c-Jun and Sp1 binding on the AP1 and Sp1 websites respectively. Promoter mutagenesis and transient transfection analyses mixed with Sp1 and c-Jun/c-Fos over-expression research point out that each Sp1 and c-Jun are required for maximal promoter exercise and, due to this fact, could positively regulate transcription of myometrial Cx43 through the initiation of labour.