What is the CpG Island importance for Huntington disease?
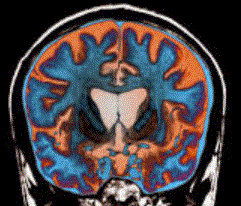
CpG islands located within the promoter region of a gene increase the probability that genes in these genomic regions are deregulated in HD. Historical studies suggested an ability to bind ssDNA reflecting a function in DNA repair and/or transcription ( Tan and Manley 2009). DNA FUS-responsive components were afterwards found enriched within promoter regions of genes shown to respond transcriptionally to FUS levels ( Tan et al.. 2012). One such gene biologically because of its connection to the neuroregressive disease Rett syndrome, was discovered to be misplaced in cells although not wild-type, FUS ( Coady and Manley 2015).
Interestingly, the splice isoform of MECP2 promoted by mutant FUS was found to have improved RNA stability yet express protein levels that were lower. N-terminal fragments of mutant huntingtin form intranuclear aggregates a hallmark that is found in a number of other polyglutamine diseases. Recent studies indicate that nuclear polyglutamine inclusions recruit transcription factors and that this recruitment influences gene expression.

Intranuclear huntingtin alters the expression of a range of genes in HD cells ( 22) and in transgenic animals ( 3 , 26 ). The atomic effect of mutant huntingtin could stem from its own interactions with a range of transcription factors, such as the nuclear receptor corepressor (N-CoR) ( 2), cyclic AMP-responsive element-binding protein (CREB)-binding protein (CBP) ( 18 , 29, 38 , 39 ), and TATA-binding proteins (TBP) ( 14, 30 ). It has been discovered that, of the transcription factors, CBP and TBP are recruited by polyglutamine inclusions ( 14, 29, 30 , 39 ). Many studies have suggested that polyglutamine inclusions are not correlated with neurodegeneration ( 19, 36 ). Therefore, recruitment of transcription factors by nuclear inclusions’ role remains to be described.
By interrogating datasets analyzed for transcriptomic regulation and existence of SNPs related to mHTT in specific genomic regions known to be involved with Type 1 and Type 2 Diabetes, we discovered that genes from the MHC/HLA locus, regarded as T1D markers 39, were down-regulated in cells expressing mHTT from the HD iPSC Consortium dataset (HLA-B).
MHC/HLA genes are responsible for the demonstration of processed antigens into T-cells, and also the activation of adaptive immune response, as the gene expression is switched on following a virus or bacterial infection or tumor mutation of proteins 40.
Sortilin SORL1 and many tetraspanins are modulated by HD from the HD iPSC Consortium dataset. An altered speed of transcription is in agreement with the hypothesis that mutant huntingtin exerts its effects by altering transcription factor action.
Comparative analysis of the promoter regions of the mouse and human CB1 genes revealed that there were a number of transcription factor binding sites which have been preserved between the two species, indicating that some shared individual factor or groups of variables could be affected by the expression of mutant huntingtin in mice and humans. Such uncertainty of the CAG repeats has also been noted in the exact same patient’s mind and sperm cells, leading to mosaicism. These number of CAG repeats seem to cause either’gain-of-function’ or loss of function of their wild-type HTT with toxic effects, such as HD-related cardiac dysfunction and muscle wasting. The majority of the mutations in muHTT disrupt its usual function, and promote pathological interactions leading to loss and dysfunction in the striatum, cortex and other parts of the mind. Thus, mutHTT interferes with several intracellular activities through aberrant interactions in addition to the accumulation of mutHTT aggregates, especially in the cell nucleus and neurophil of those affected neurons, finally interrupting several cell processes, including protein degradation, mitochondrial respiration and transcription, resulting in cognitive malfunction as well as cell death. Apparently the growth in CAG repeats are often as high as 1,000 in subsets of neurons while the increase is lower. Recent genome-wide single nucleotide polymorphism (SNP) association studies showed that MLH1 (MutL homolog 1, a DNA mismatch repair gene) and a SNP within a nuclear factor-κB binding site in the HTT promoter can play a part in the modified onset of HD.
To determine the gene disrupted in those stretch mutants we sequenced the genomic DNA flanking a few stretch P-element alleles and discovered that these P-element mutations were inserted in a novel Drosophila gene that covered >115 kb of genomic DNA.
This single gene had been annotated as three separate genes and given that the titles CG12418, CG12802, and CG33975 (previously CG11676 and CG32469). Our in-depth analysis of ESTs and cDNAs in that area of the genome revealed at least four different transcripts sharing overlapping exons.
SLC2A4RG was initially characterized and named for its ability to bind to the enhancer of the Glucose Transporter 4 gene ( Oshel et al.. 2000 ). Glut4EF proteins can also be highly similar to papillomavirus binding factor (PBF) ( Boeckle et al.. 2002 ), also referred to as HDBP2 ( Tanaka et al.. 2004), and ZNF395 ( Stoeckman et al.. 2006 ) and in our quest of the database we discovered a previously uncharacterized human EST ZNF704 like the mouse EST Zfp704 ( Blackshaw et al.. 2004), also called mouse glucocorticoid-induced receptor 1 (Gig) that has been highly related to Glut4EF proteins also signifies a third mammalian relative. Therefore, the gene disrupted in elongate mutants signifies the Drosophila member of a family of transcription factors. Our results add to the growing body of understanding of sortilins as proteins that influence processing of antigens in autoimmunity and other purposes related to the immune system by providing evidence that gene expression or genetic diversity of sortilin and MHC/HLA genes have been associated with mHTT in HD.
Relationship with Alzheimer Disease
These findings contribute to an improved understanding regarding the control of traffic of sugar receptors, insulin and Alzheimer’s Disease proteins in the trans-Golgi network, the endosome and the plasma-membrane in normal and HD states, using versions of HD described within this report.
We revealed that mHTT is associated with enhanced protein expression of sortilin SORCS1 in ST14A cells, in agreement with our previous report for mRNA atoms of SORCS1 and there exists allele association (also referred to as Linkage Disequilibrium) of regions near SORT1, SORL1, SORCS1, SORCS2 and SORCS3 with receptor variants in the region of HTT gene specifically in human HD instances.
Eleven sortilins SNPs previously detected by Reitz et al. (2013), who investigated the participation of SORCS1 variants with AD, were detected in our analysis.
It is important to contrast the number of people involved in many GWAS studies into the amount of HD cases reported here (Reitz 2013 individuals: n = 11,840 instances and 10,931 controls; Labadorf dataset 2015: n = 20 instances and 49 controls).
