The mechanisms for the regulation of homeotic genes are poorly understood in most organisms, together with vegetation. We recognized BASIC PENTACYSTEINE1 (BPC1) as a regulator of the homeotic Arabidopsis thaliana gene SEEDSTICK (STK), which controls ovule id, and characterised its mechanism of motion.
A mixture of tethered particle movement evaluation and electromobility shift assays revealed that BPC1 is ready to induce conformational adjustments by cooperative binding to purine-rich components current within the STK regulatory sequence.
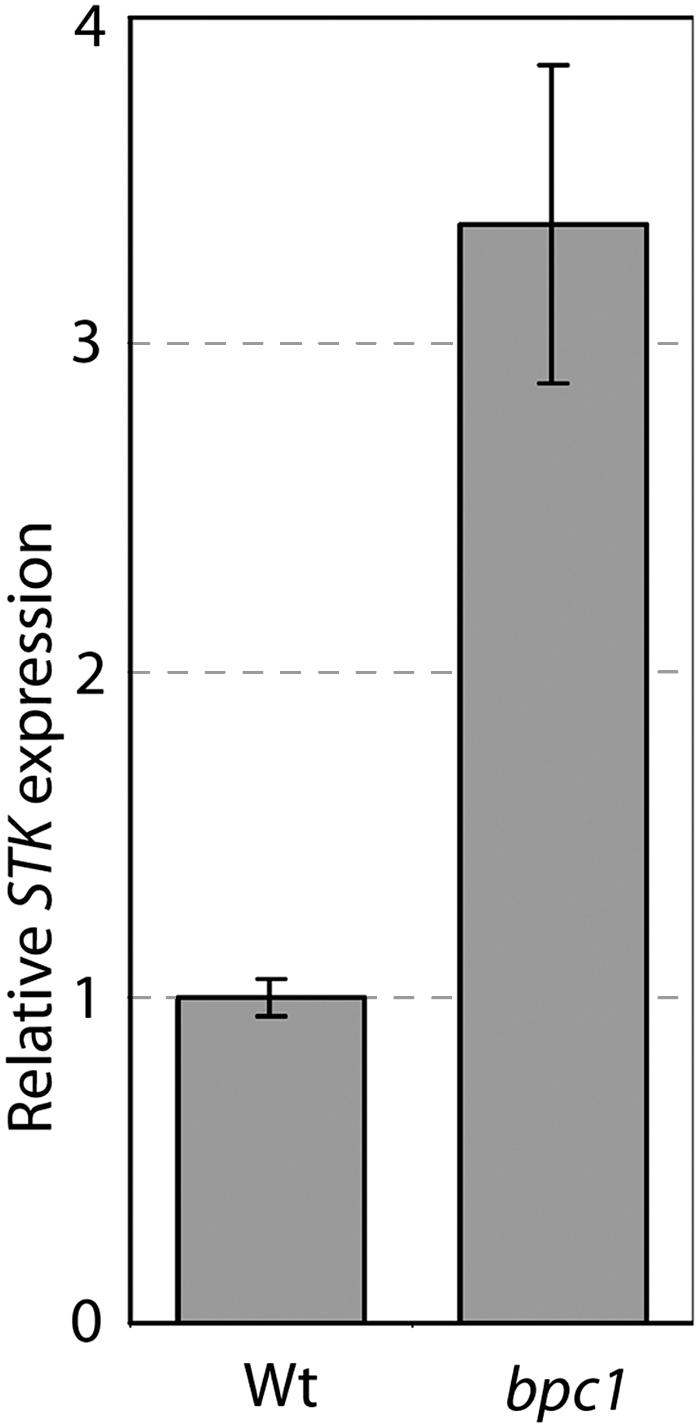
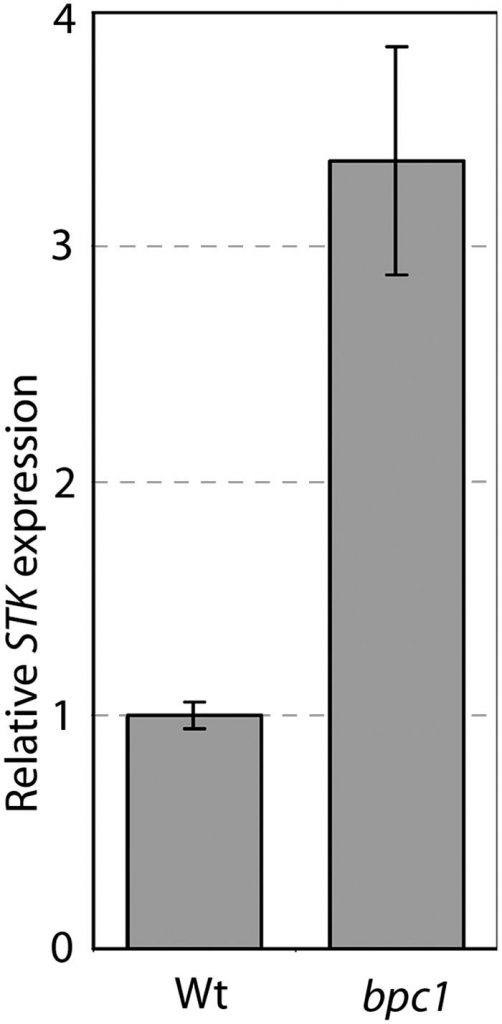
Evaluation of STK expression within the bpc1 mutant confirmed that STK is upregulated. Our outcomes give perception into the regulation of gene expression in vegetation and supply the premise for additional research to grasp the mechanisms that management ovule id in Arabidopsis.
Particular binding of the regulatory protein ExpG to promoter areas of the galactoglucan biosynthesis gene cluster of Sinorhizobium meliloti–a mixed molecular biology and drive spectroscopy investigation.
Particular protein-DNA interplay is prime for all points of gene transcription. We deal with a regulatory DNA-binding protein within the Gram-negative soil bacterium Sinorhizobium meliloti 2011, which is able to fixing molecular nitrogen in a symbiotic interplay with alfalfa vegetation.
The ExpG protein performs a central function in regulation of the biosynthesis of the exopolysaccharide galactoglucan, which promotes the institution of symbiosis. ExpG is a transcriptional activator of exp gene expression.
We investigated the molecular mechanism of binding of ExpG to 3 related goal sequences within the exp gene cluster with normal biochemical strategies and single molecule drive spectroscopy based mostly on the atomic drive microscope (AFM).
Binding of ExpG to expA1, expG-expD1, and expE1 promoter fragments in a sequence particular method was demonstrated, and a 28 bp conserved area was discovered. AFM drive spectroscopy experiments confirmed the particular binding of ExpG to the promoter areas, with unbinding forces starting from 50 to 165 pN in a logarithmic dependence from the loading charges of 70-79000 pN/s.
Two totally different regimes of loading rate-dependent behaviour have been recognized. Thermal off-rates within the vary of okay(off)=(1.2+/-1.0) x 10(-3)s(-1) have been derived from the decrease loading price regime for all promoter areas. Within the higher loading price regime, nonetheless, these fragments exhibited distinct variations that are attributed to the molecular binding mechanism.